Basic methods for class SansSouci
nHyp: get the number of hypotheses
nObs Get the number of observations
label Get the label of a post hoc method
Print 'SansSouci' objects
pValues: get p-values
thresholds: get thresholds
Usage
# S3 method for class 'SansSouci'
nHyp(object)
# S3 method for class 'SansSouci'
nObs(object)
# S3 method for class 'SansSouci'
label(object)
# S3 method for class 'SansSouci'
print(x, ..., verbose = FALSE)
# S3 method for class 'SansSouci'
pValues(object)
# S3 method for class 'SansSouci'
foldChanges(object)
# S3 method for class 'SansSouci'
thresholds(object)Examples
data(expr_ALL, package = "sanssouci.data")
groups <- ifelse(colnames(expr_ALL) == "NEG", 0, 1)
table(groups)
#> groups
#> 0 1
#> 42 37
a <- SansSouci(Y = expr_ALL, groups = groups)
print(a)
#> 'SansSouci' object:
#> Number of hypotheses: 9038
#> Number of observations: 79
#> 2 samples
#>
nHyp(a)
#> [1] 9038
nObs(a)
#> [1] 79
label(a)
#> NULL
res <- fit(a, B = 100, alpha = 0.1)
label(res)
#> [1] "Simes"
print(res)
#> 'SansSouci' object:
#> Number of hypotheses: 9038
#> Number of observations: 79
#> 2 samples
#>
#> Parameters:
#> Test function: rowWelchTests
#> Number of permutations: B=100
#> Significance level: alpha=0.1
#> Reference family: Simes
#> (of size: K=9038)
#>
#> Output:
#> Calibration parameter: lambda=0.1210904
str(pValues(res))
#> num [1, 1:9038] 0.9647 0.5203 0.9516 0.8438 0.0045 ...
#> - attr(*, "dimnames")=List of 2
#> ..$ : chr "Group 1 vs group 0"
#> ..$ : NULL
str(foldChanges(res))
#> num [1, 1:9038] -0.0044 0.09394 -0.00426 -0.00886 0.27775 ...
#> - attr(*, "dimnames")=List of 2
#> ..$ : chr "Group 1 vs group 0"
#> ..$ : NULL
str(thresholds(res))
#> num [1:9038] 1.34e-05 2.68e-05 4.02e-05 5.36e-05 6.70e-05 ...
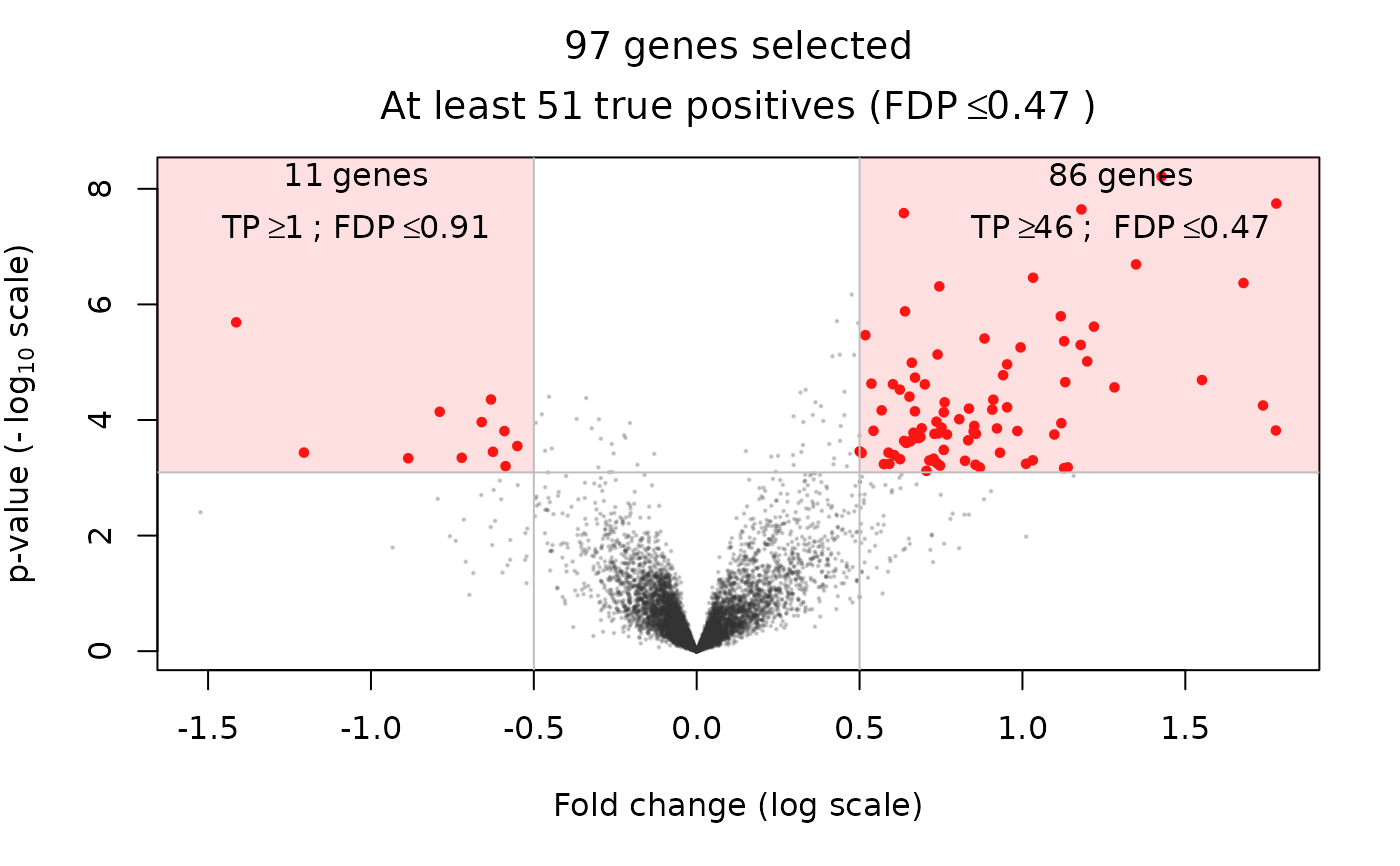
volcanoPlot(res, q = 0.05, r = 0.5)