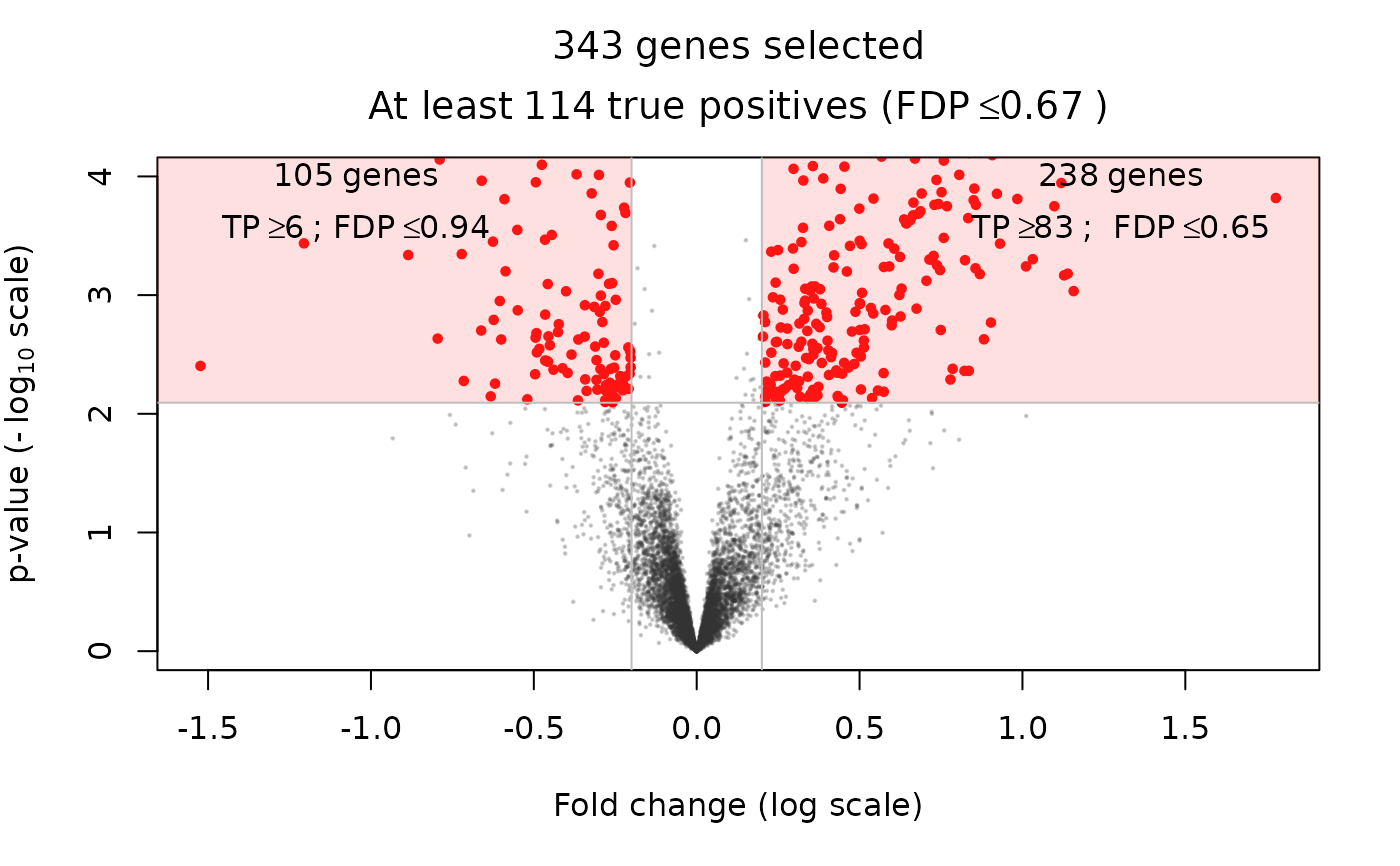
Volcano plot
Usage
volcanoPlot(x, ...)
# S3 method for class 'SansSouci'
volcanoPlot(
x,
fold_changes = foldChanges(x)[contrast_name, ],
p_values = pValues(x)[contrast_name, ],
p = 1,
q = 1,
r = 0,
contrast_name = x$input$contrast_name[1],
cex = c(0.2, 0.6),
col = c("#33333333", "#FF0000", "#FF666633"),
pch = 19,
ylim = NULL,
...
)Arguments
- x
An object of class
SansSouci- ...
Other arguments to be passed to volcanoPlot.numeric
- fold_changes
An optional vector of fold changes, of the same length as
nHyp(object), use for volcanoPlot x-axis. If not specified,- p_values
A vector of p-values, of the same length as
nHyp(object), use for volcanoPlot x-axis- p
A numeric value, the p-value threshold under which genes are selected
- q
A numeric value, the q-value (or FDR-adjusted p-value) threshold under which genes are selected
- r
A numeric value, the absolute fold change above which genes are selected
- contrast_name
A character value, the selected contrast. Should be chosen in
x$input$contrast_name.- cex
A numeric vector of length 2, the relative magnification factor for unselected (
cex[1]) and unselected (cex[2]) genes.- col
A vector of length 3
- pch
An integer or single character string specifying the plotting character, see par
- ylim
A numeric vector of length 2, the
ylimits of the plot